Analyzing Single Nucleotide Polymorphisms and Their Chromosomal Distribution to Unravel the Genetic Landscape of Melanoma
Analyzing Single Nucleotide Polymorphisms and Their Chromosomal Distribution to Unravel the Genetic Landscape of Melanoma
Angelina Huang *1, Dr. Robert Aguilar 2
*Correspondence to: Angelina Huang,.
Copyright
© 2024 Angelina Huang. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 16 July 2024
Published: 01 August 2024
Abstract
Melanoma, a malignant skin cancer, continues to pose significant challenges in early detection, optimal treatment strategies, and understanding its etiology. This study aimed to elucidate the complex genetic landscape of melanoma by analyzing 2,641 Single Nucleotide Polymorphisms (SNPs) across 18 genomic variant categories within a melanoma cohort. The analysis revealed diverse genetic variations, including Missense and Frameshift Variants, with a significant number of unknown variants that require further exploration. A significant association (p < 0.05) was found between the identified variants and the melanoma cohort. The study's findings underscore the heterogeneous nature of melanoma and emphasize the potential need for personalized therapeutic approaches. This comprehensive study contributes valuable insights into melanoma's genetic landscape and holds promise for future interventions and personalized prevention and treatment strategies.
Keywords: Melanoma, Single Nucleotide Polymorphisms (SNPs), Genomic Variants, Personalized Medicine, Therapeutic Targets, Early Detection.
Analyzing Single Nucleotide Polymorphisms and Their Chromosomal Distribution to Unravel the Genetic Landscape of Melanoma
Introduction
Cancer
Melanoma, a neoplasm arising from melanin-forming cells, is a primary malignancy that typically appears on the skin but can also occur in the eyes, and mouth (Mayo Foundation for Medical Education and Research, 2023). As a common form of skin cancer, melanoma impacts an estimated 97,000 individuals in the United States annually, culminating in approximately 8,000 fatalities each year (American Cancer Society, 2023). The disease's etiology is multifaceted, with critical contributing factors comprising race, regional sun exposure, and age. Remarkably, white individuals exhibit a risk of developing melanoma that is 20 times greater than that of other racial groups.
Furthermore, men over the age of 65 are twice as likely as women to receive a melanoma diagnosis. Additionally, regions with elevated levels of sun exposure, such as Australia, have been found to have the highest incidence rates of melanoma globally (American Academy of Dermatology Association 2023).
The first known description of melanoma can be traced back to the ancient writings of Hippocrates, indicating the disease's long-standing presence throughout human history (Morton et al. 2003). In modern society, its prevalence is especially prominent among young adults, particularly women, making it the most common form of cancer in this demographic (American Academy of Dermatology Association 2023). The societal and economic implications of this widespread disease are profound: Each year in the United States, an estimated 5 million people receive treatment for melanoma, translating to a staggering annual cost of $8.1 billion (Skin Cancer Foundation, 2023). These figures underline the imperative need for ongoing research into prevention, early detection, and efficient treatment strategies to mitigate the burden of this prevalent and life-threatening condition.
Stages
Melanoma is frequently identified through the observation of progressively pigmented, anomalous cutaneous lesions. To effectively discern malignant melanocytic lesions from benign dermatological conditions, healthcare practitioners often employ the "ABCDE" mnemonic: asymmetry, border irregularity, color variation, diameter enlargement, and evolution over time (CDC, 2023). Melanoma is frequently identified through the observation of progressively pigmented, anomalous cutaneous lesions. To effectively discern malignant melanocytic lesions from benign dermatological conditions, healthcare practitioners often employ the "ABCDE" mnemonic: asymmetry, border irregularity, color variation, diameter enlargement, and evolution over time (CDC, 2023).
Understanding the characteristics of suspicious lesions is merely the first step in a multifaceted diagnostic and treatment process. Transitioning from lesion identification, the next critical component in managing melanoma involves staging the disease. With each stage bearing significant implications for patient prognosis and treatment strategy, melanoma's progression can be delineated into four stages that denote the extent and severity of the disease (see Figure 1 below). Stage 0 melanoma, commonly referred to as melanoma in situ, is marked by neoplastic growth confined exclusively to the epidermis, the skin's outermost layer. In this initial stage, the abnormal cells have not yet penetrated deeper layers, such as the dermis or subcutaneous tissue, indicating that the disease has not spread beyond its point of origin. In Stage 1, the malignant melanocytes have penetrated beyond the skin's outermost layer into the underlying dermis. This stage is characterized by increased tumor thickness compared to Stage 0, attributable to the vertical growth phase behavior of the neoplastic cells.
FIGURE 1: Progression of Melanoma Stages. Melanoma undergoes staging ranging from 0 to IV, capturing its evolving severity. Stage 0 illustrates in situ cancer, confined to the epidermis. Advancing to Stage 1, the cancer infiltrates the dermis. In Stage 2, the cancer deepens and may show ulceration. Stage 3 marks metastasis, with melanoma extending to lymph nodes. The most advanced, Stage 4, portrays melanoma's dissemination to adjacent organs such as the brain, lungs, and liver (Source: SITC).
Stage 2 is identified by continued confinement of the malignancy to the dermis, however, the defining features of this stage include the increased tumor thickness and the presence of ulceration (an erosive process that disrupts the epidermis), creating an open wound on the skin surface. Given the melanoma's deeper infiltration in this stage, the probability of metastasis, i.e., the dissemination of cancerous cells to disparate regions of the body, is significantly heightened (Aim at Melanoma, 2023). In Stage 3, the malignancy has extended to the regional lymph nodes which signifies a more aggressive disease course and mandates more immediate therapeutic intervention.
Stage 4 melanoma exemplifies the metastatic dissemination extending beyond its initial site to distant organs. As depicted in the lower-right corner of Figure 1, this may encompass crucial organs such as the brain, liver, and lungs, marking a pivotal milestone in the progression of the disease (Aim at Melanoma, 2023). Typically linked with an unfavorable prognosis, this stage demands a comprehensive, multifaceted systemic therapeutic strategy.
Treatment
The management strategies of melanoma vary in severity, encompassing a wide spectrum of treatment from local surgical interventions to more advanced systemic modalities. At the initial stages (0-2), surgical excision is primarily employed; this involves the resection of the neoplastic growth along with an appropriate margin of the surrounding healthy dermis (UPenn Medicine, 2023). Upon progression to stages 2-3, when the malignancy becomes more aggressive, consideration is given to immunotherapy. This methodology hinges on the principle of augmenting innate immunological mechanisms to effectively counter neoplastic cells. Therapeutic agents are typically administered intravenously, specifically engineered to target proteins expressed on the neoplastic cells.
There are several subdivisions of immunotherapy including, but not limited to, immune checkpoint inhibitors, interleukin-based therapies, and oncolytic virotherapy (UPenn Medicine, 2023).
Immune checkpoint inhibitors augment the cytotoxic potential of white blood cells by targeting and neutralizing inhibitory proteins that otherwise impede these immune cells from effectively eradicating the malignant entities (National Cancer Institute 2022). On the other hand, interleukin immunotherapy involves the administration of interleukins, essential modulators of white blood cell activity, thereby attenuating the growth and proliferation of melanoma tumors. Oncolytic virotherapy encompasses the utilization of genetically modified viruses, selectively designed to target and eliminate neoplastic cells. These immunotherapeutic interventions are not solely restricted to active stages of the malignancy but are also instrumental in maintaining immunological robustness during the convalescent phase post-melanoma treatment (UPenn Medicine, 2023). Targeted molecular therapy has emerged as a formidable contender in the therapeutic armamentarium against melanoma. One such strategy involves the inhibition of the mutated BRAF gene, a frequent genetic aberration in melanoma that precipitates anomalous and unregulated cellular proliferation. Lastly, radiotherapy remains an indispensable tool in managing melanoma. This modality operates by delivering high-energy radiation, specifically directed towards the loco-regional tumors, effectively compromising the DNA integrity within the malignant cells, and precluding further propagation of the disease (Aim at Melanoma, 2023).
Therapeutic approaches toward melanoma remain an enigmatic challenge. Current cancer treatments suffer from significant drawbacks, manifesting as a host of adverse effects. These can range from gastrointestinal disturbances such as nausea to hematological complications, compromised immunity, and an increased predilection for other illnesses (MSK, 2022). Even in the aftermath of successful treatment, the shadow of recurrence looms, with melanoma documented to re-emerge anywhere between 5 and 25 years post-treatment (American Academy of Dermatology, 2023). Another formidable barrier lies in the socioeconomic realm: the costs of melanoma treatments, which vary greatly depending on the stage of the disease, can be prohibitive. Treatment expenses can range from $400 for early-stage melanoma to an astronomical $50,000 for advanced stages (Menchaca, 2023). This significant financial burden further compounds the issue, making access to potentially life-saving treatments challenging for many individuals.
Gene
The CDKN2A gene is intricately linked to melanoma, with mutations in this gene accounting for approximately 40% of melanoma cases and representing a significant familial predisposition to this form of skin cancer (Rossi et al., 2019). It serves as a vital source of instructions for synthesizing the p16 and p14 proteins. Both p16 and p14 proteins function as tumor suppressors, orchestrating cellular processes within distinct phases of the cell cycle. As illustrated in Figure 2, p16 plays a pivotal role in cell cycle deceleration by inhibiting cyclin-dependent kinases (CDKs), imperative protein families that govern central functions such as transcription and mRNA processing (Liu et al., 2012). Similarly, the p14 protein's mechanism involves activating p53, a transcription factor crucial for maintaining genomic integrity. Situated on the 9th chromosome, the CDKN2A gene is frequently prone to mutations that bear connections to cancer. These variants can manifest as either germline mutations, inherent genetic alterations, or somatic mutations, which arise during early or adult cell development (Liu et al., 2012).
FIGURE 2: CDKN2A's Cell Cycle Regulation. CDKN2A's genetic products, p14, and p16, exert pivotal control over the cell cycle. Through p14's suppression of MDM2, p53 activity is enhanced, curtailing unchecked DNA division. Simultaneously, p16 acts as a brake on cyclin-dependent kinases (CDKs), halting progression from G2 phase to metaphase. This intricate interplay ensures regulated cell growth and genomic stability (Research Gate, 2014).
SNPs
Single Nucleotide Polymorphisms (SNPs) are genetic variations involving a single base pair in DNA, occurring at a frequency of over 1% in the population. These variants are frequently situated in non-coding regions of the genome, encompassing diverse mRNA locales such as promoters, exons, and introns, as well as the 5' and 3' untranslated regions (UTRs) of genes (Deng et al., 2017). Their impact can reverberate across multiple biological processes, potentially leading to alterations in amino acid composition, disturbances in the cell cycle, or the perturbation of gene expression control (Shastry, 2009). Significantly, SNPs serve as valuable "biological markers" that facilitate the identification of gene variations, enabling the establishment of connections between these genetic variations and the onset of diseases.
In the context of melanoma, SNPs have emerged as important factors that contribute to disease susceptibility and progression. Research has unveiled associations between specific SNPs and melanoma risk, shedding light on the underlying genetic factors that could predispose individuals to this malignancy. These variations have been identified in genes linked to crucial processes such as DNA repair, cell cycle regulation, and immune response. For instance, SNPs in the CDKN2A gene have been linked to increased susceptibility to this skin cancer (Gandini et al., 2005). Similarly, SNPs in genes involved in pigmentation, immune response modulation, and DNA repair pathways have been implicated in altering melanoma risk and clinical outcomes (Cust et al., 2011; Han et al., 2008). As we deepen our understanding of these genetic variations and their functional implications, the potential to harness this knowledge for personalized prevention and treatment strategies becomes increasingly promising.
Methodology
In the present study, we conducted an exhaustive analysis of sequence reads derived from melanoma patients as well as control subjects to identify genomic variants and scrutinize their possible implications. The reference genome for Homo sapiens chromosome 9, obtained from Ensembl (Release 104) (Cunningham et al., 2021), served as a benchmark for precise juxtaposition and interpretation of genomic alterations recognized in our investigation. The analytical pipeline we implemented incorporated a selection of Sequence Read Archive (SRA) sequences contingent upon library construction strategy and experimental design. We procured these SRA files utilizing the fastq-dump utility, part of the SRA Toolkit v2.10.7 (NCBI, 2021), with subsequent computations executed via the command-line interface. An initial appraisal of sequence read quality was facilitated through FastQC (v0.11.9) (Andrews, 2010).
For sequence trimming, we employed Trimmomatic (v0.39) (Bolger et al., 2014), concentrating specifically on regions demonstrating compromised base quality, indicated by a Phred quality score below 33, as deciphered from FastQC-generated HTML reports. This process culminated in the enhancement of overall sequence data quality through the excision of low-quality bases.
We utilized Bowtie2 (v2.4.2) (Langmead & Salzberg, 2012) for the alignment of sequences to the reference human genome and for indexing of the reads. The resultant Sequence Alignment/Map (SAM) file was converted into a Binary Alignment/Map (BAM) file utilizing SAMtools (v1.11) (Li et al., 2009), a comprehensive suite designed for manipulating high-throughput sequencing data. Following coordinate-based sorting of the BAM file, the coverage depth at each genomic position was ascertained using the SAMtools 'depth' command, thereby highlighting regions prone to encompass genetic variants through the depth of coverage analysis across the relevant chromosome.
SNPs were pinpointed employing the binary variant call format (BCF) utilities from the SAMtools suite. The derived BCF file underwent stringent filtering through BCFTools (v1.11), retaining solely high-confidence variants. This robust process ensured the inclusion of only reliable and insightful SNPs in our analysis. We further validated these SNPs by cross-referencing our findings with a reference dataset comprising SNPs from a healthy population, leveraging the Pandas library, a powerful tool for data manipulation in Python. This procedure facilitated the elimination of any matching SNPs from our dataset, thereby enabling our focus on melanoma-specific genomic alterations.
Statistical analyses were executed to assess the association between identified variants within the melanoma and control cohorts. Chi-square tests were utilized for the evaluation of categorical variables. A contingency table was constructed from the CSV file, exhibiting the counts of subjects with and without specific variants across both cohorts. The Python SciPy library was utilized to compute the chi-square statistic and the corresponding p-value. The results unveiled a significant association (p < 0.05) between the identified variants and the melanoma cohort, offering persuasive evidence to reject the null hypothesis.
We further associated SNP accession numbers with their genomic consequences utilizing a Python script that we developed. SNP accession numbers were extracted from the SAMtools-generated BCF file, and the Ensembl REST API was employed to retrieve pertinent information regarding each SNP, including genomic location, allelic configuration, and genomic consequences. The parsed JSON responses facilitated the extraction of relevant data, which was subsequently merged with the associated genomic consequences and stored in a CSV file for later analysis. This methodology permitted the efficient extraction of information regarding the genomic consequences of each SNP, essential for understanding their potential influence on health outcomes. The deployment of a Python script facilitated automation, considerably mitigating the risk of manual data entry errors. The Python scripts deployed in this study, inclusive of the script for associating SNP accession numbers with their genomic consequences, are publicly available on our GitHub repository
(https://github.com/crisprmax/SNP-identifier-Python). This repository provides access to the code implemented in this study, accompanied by comprehensive documentation and user guidelines for the execution of the scripts.
Results
Our investigation into the sequence reads, derived from melanoma patients and control subjects, represents an essential step in our ongoing efforts to understand the genetic underpinnings of this complex disease. By analyzing hundreds of genomic sequences, we have uncovered significant insights into the specific genomic variants associated with melanoma. This detailed examination has not only revealed previously unrecognized connections between particular genetic variations and malignancy but also sheds light on their possible functional implications. This study extends our understanding of how these genomic variants may provide critical insights that could inform future diagnostic approaches, targeted treatments, and personalized care strategies for individuals affected by this life-threatening condition.
Distribution and Categories of SNPs
Our analysis uncovered a diverse array of 2,641 SNPs across 18 distinct genomic variant categories within the melanoma cohort. These SNPs spanned a spectrum of genetic variations, from those with unknown effects, requiring further exploration (1420 SNPs), to specific, well-defined alterations such as Missense (24 SNPs) and Frameshift Variants (3 SNPs). The category labeled as 'unknown effects' represents a significant portion of these variations, suggesting potential novel pathways and mechanisms of melanoma development. The Missense and Frameshift Variants, although small in number, are of particular interest as they may directly affect protein structure and function. These variants can result in the substitution of one amino acid for another or alterations in the reading frame, leading to profound biological consequences. This understanding of the genetic variations provides a window into the complexity and heterogeneity inherent in melanoma's genetic landscape (Table 1).
Table 1: Summary of SNPs Identified in a Melanoma Cohort. The table provides an overview of the different categories of SNPs, including their count and a brief description of their potential effects or characteristics (Strachan & Read, 2015).
Figure 3 below showcases the genomic variants identified as statistically significant in our study, with a p-value of less than 0.05, while excluding those classified as 'unknown.'
Figure 3: Distribution of SNPs Across Different Genomic Variant Categories in a Melanoma Cohort. This bar graph illustrates the diversity and frequency of 9 distinct categories of SNPs detected in a melanoma cohort. The x-axis categorizes the genomic variants, encompassing a wide spectrum from Intron variants to specific alterations like Missense and 3’UTR Variants.
Significant Association with Melanoma
Our chi-square tests revealed a significant association (p < 0.05) between the identified variants and the melanoma cohort. This finding provides persuasive evidence of the role these variants may play in melanoma susceptibility, progression, and clinical outcomes, strengthening our understanding of the genomic factors that influence this malignancy.
Genomic Consequences of Identified SNPs
The genomic consequences of each SNP were successfully associated with their accession numbers. With the use of the Ensembl REST API, we were able to extract pertinent information regarding the genomic location, allelic configuration, and genomic consequences. This information is vital for understanding the potential influence of these SNPs on health outcomes, further enriching our perspective on the genetic variations in melanoma. The extensive diversity and complexity of these genetic variations highlight the multifaceted nature of melanoma. This intricate genetic profile emphasizes the potential need for personalized therapeutic approaches, reinforcing the importance of continued research and development in genomic analysis techniques.
Chromosomal Distribution of Unique SNPs
Figure 4 offers an intricate visualization that maps the spatial distribution of unique SNPs across designated loci on Chromosome 9, a chromosome of particular interest in melanoma research (Canon et al., 2013). In this illustration, genes meeting or exceeding a threshold of 18 unique SNPs are clearly highlighted and labeled. This method of visual representation serves to accentuate chromosomal regions and specific genes that may have a significant association with melanoma. The clustering of unique SNPs within distinct areas of Chromosome 9 suggests the existence of genomic hotspots where genetic alterations are especially concentrated. These hotspots provide valuable insights into potential mechanisms contributing to the initiation and progression of melanoma.
Figure 4: The distribution of unique SNPs across specific positions on Chromosome 9. Each black circle in the plot corresponds to a specific gene, with the position on the x-axis (represented as a tic mark) reflecting the gene's location on the chromosome, and the y-axis indicating the total count of unique SNPs for that gene. Genes that meet or exceed a threshold of 18 unique SNPs are highlighted with white circles and labeled with their corresponding gene names (p < 0.05).
Summary of Major Findings
The major findings of our study encompass several critical areas of interest in the genetic landscape of melanoma. A substantial number of the identified SNPs (1420) fell into the Unknown category, emphasizing the need for further research to elucidate their potential effects on melanoma development and progression. The prevalence of Non-Coding Transcript Exon Variants (678) and Intron Variants (280) suggests potential influences on gene regulation, RNA stability, expression, or splicing that may be relevant to melanoma. Specific alterations, such as Missense and Frameshift Variants, were identified, indicating potential direct effects on protein structure and function. Furthermore, the spatial distribution of SNPs on Chromosome 9, as detailed in our earlier section, provides targeted areas for further investigation, potentially uncovering novel genes or pathways associated with melanoma risk. These insights collectively contribute to a multifaceted understanding of the genetic variables influencing this complex and often fatal disease.
Discussion
This study of melanoma's genetic landscape has unearthed several critical insights, chief among them the identification of 2,641 SNPs with significant association with melanoma susceptibility and progression. Of particular interest are the chromosomal insights provided by Figure 4, which delineate potential therapeutic targets, shedding light on specific genomic regions and their role in melanoma. The findings reveal a complex distribution of genetic variants, including Missense and Frameshift Variants, consistent with the recognized heterogeneous nature of melanoma. The extensive presence of unknown variants underscores the uncharted territories within melanoma genetics, necessitating further exploration.
These findings carry far-reaching implications for both the scientific community and personalized medicine, opening avenues for more precise and targeted therapeutic strategies. Understanding the chromosomal distribution enriches our comprehension of melanoma's genetic complexity, enhancing prospects for diagnosis and treatment. Despite its promise, the study has limitations, particularly the substantial number of unknown categorized SNPs and potential restrictions in the sample's generalizability. These factors beckon further investigation.
Future research endeavors should prioritize the functional analysis of these unknown variants and extend the findings to diverse patient populations. This could facilitate novel therapeutic interventions, particularly by delving into the specific genomic regions highlighted in Figure 4. Our results open up a promising pathway towards personalized prevention and innovative treatment modalities for melanoma, providing a fresh perspective on this aggressive cancer form. However, consideration of alternative explanations, such as epigenetic or environmental influences, should remain central in subsequent research to present a multifaceted understanding.
The study fittingly aligns with the broader paradigm of personalized medicine, challenging conventional, one-size-fits-all approaches, and emphasizes the primacy of genomic information in tailoring interventions.
Clinically, these findings harbor the potential to transform practice by enabling more targeted screening, early detection, and intervention, leading to personalized treatment plans and improved patient outcomes. Ultimately, the novelty of our approach, particularly in exploring unknown variants and their specific chromosomal distribution, fills a crucial knowledge gap in melanoma research. It provides an innovative lens through which to view the field, laying the groundwork for more specialized and individualized therapeutic strategies for melanoma.
Conclusion
This study represents a significant advancement in our understanding of the genetic landscape of melanoma. By identifying and categorizing 2,641 SNPs and exploring their chromosomal distribution, we have highlighted critical areas of interest for future research and therapeutic intervention. The complexities uncovered within the genetic variants, including the prevalence of unknown categories, echo the intricate and heterogeneous nature of melanoma, which demands a tailored and patient-centric approach to diagnosis and treatment.
The associations found between the identified variants and melanoma susceptibility provide compelling evidence of the genetic factors' role in this aggressive form of cancer. While this study opens up promising avenues for personalized medicine, it also brings to light the challenges and limitations that need to be addressed in future research. Specifically, the elucidation of unknown variants and the consideration of broader patient populations are crucial steps toward more effective prevention and targeted treatment strategies. This research represents an encouraging step forward in the ongoing quest to understand, diagnose, and effectively treat melanoma.
References
1. Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc
2. Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data.
3. Bioinformatics, 30(15), 2114-2120. https://doi.org/10.1093/bioinformatics/btu170
4. Cannon-Albright, L. A., Teerlink, C. C., Farnham, J. M., Thomas, A. W., Zone, J. J., & Leachman, S. A. (2013).
5. Linkage analysis of extended high-risk pedigrees replicates a cutaneous malignant melanoma predisposition locus on chromosome 9q21. The Journal of investigative dermatology, 133(1), 128–134. https://doi.org/10.1038/jid.2012.271
6. Centers for Disease Control and Prevention. (2023, April 18). What are the symptoms of skin cancer?. Centers for Disease Control and Prevention. https://www.cdc.gov/cancer/skin/basic_info/symptoms.htm#:~:text=Border%3A%20Is%20the%20border% 20irregular,past%20few%20weeks%20or%20months%3F
7. Cunningham, F., Achuthan, P., Akanni, W., Allen, J., Amode, M. R., Armean, I. M., ... & Flicek, P. (2021). Ensembl 2021. Nucleic Acids Research, 49(D1), D884-D891. https://doi.org/10.1093/nar/gkaa942
8. Cust, A. E., Goumas, C., Vuong, K., Davies, J. R., Barrett, J. H., Holland, E. A., Schmid, H., Agha-Hamilton, C., Armstrong, B. K., Kefford, R. F., Mann, G. J., & Investigators, G. (2011). MC1R genotype as a predictor of early-onset melanoma, compared with self-reported and physician-measured traditional risk factors: An Australian case-control-family study. BMC cancer, 11, 283.
9. Deng, L., Xu, X., Li, Y., Yang, F., & Wei, Z. (2017). Exploration and practice of using exome sequencing in routine genetic diagnosis. Yi chuan= Hereditas, 39(11), 1033–1041.
10. Deng, N., Zhou, H., Fan, H., & Yuan, Y. (2017, November 7). Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. https://www.oncotarget.com/article/22372/text/#:~:text=SNPs%20are%20located%20in%20gene,the%20in dividual%20SNPs%20are%20located
11. Gandini, S., Sera, F., Cattaruzza, M. S., Pasquini, P., Abeni, D., Boyle, P., & Melchi, C. F. (2005). Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. European journal of cancer (Oxford, England: 1990), 41(1), 45–60.
12. Han, J., Kraft, P., Nan, H., Guo, Q., Chen, C., Qureshi, A., Hankinson, S. E., Hu, F. B., Duffy, D. L., Zhao, Z. Z., Martin, N. G., Montgomery, G. W., Hayward, N. K., Thomas, G., Hoover, R. N., & Chanock, S. J. (2008). A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS genetics, 4(5), e1000074.
13. Immune checkpoint inhibitors. National Cancer Institute. (n.d.). https://www.cancer.gov/about-cancer/treatment/types/immunotherapy/checkpoint-inhibitors
14. Immunotherapy for Melanoma. Pennmedicine.org. (n.d.). https://www.pennmedicine.org/cancer/types-of-cancer/melanoma/melanoma-treatments/melanoma-immuno therapy#:~:text=These%20medications%20help%20T%20cells,support%20the%20overall%20immune%2 0system.
15. Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357-359. https://doi.org/10.1038/nmeth.1923
16. Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., ... & Durbin, R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078-2079. https://doi.org/10.1093/bioinformatics/btp352
17. Liu, X.-Y., Petska, S., & Shi, Y.-F. (2012). Recent Advances in Cancer Research and Therapy. Mayo Foundation for Medical Education and Research. (2023, July 22). Melanoma. Mayo Clinic. https://www.mayoclinic.org/diseases-conditions/melanoma/symptoms-causes/syc-20374884#:~:text=Melan omas%20can%20develop%20anywhere%20on,your%20hands%20and%20fingernail%20beds.
18. Melanoma skin cancer statistics. Melanoma Skin Cancer Statistics | American Cancer Society. (n.d.). https://www.cancer.org/cancer/types/melanoma-skin-cancer/about/key-statistics.html
19. Menchaca, C. (2021, October 11). The cost of skin cancer: Diagnosis, treatment, and care. HealthMatch. https://healthmatch.io/skin-cancer/cost-of-skin-cancer-treatment
20. Morton, D. L., Essner, R., Kirkwood J. M., Wollman R. C. (2003). Home - books - NCBI. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/books
21. National Center for Biotechnology Information (NCBI). (2021). SRA Toolkit v2.10.7. Retrieved from https://github.com/ncbi/sra-tools
22. Radiation therapy for melanoma. Memorial Sloan Kettering Cancer Center. (1970, January 1). https://www.mskcc.org/cancer-care/types/melanoma/treatment/radiation-therapy-melanoma
23. Rossi, M., Pellegrini, C., Cardelli, L., Ciciarelli, V., Di Nardo, L., & Fargnoli, M. C. (2019, January 31). Familial melanoma: Diagnostic and management implications. Dermatology practical & conceptual. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6368081/#:~:text=CDKN2A%20is%20the%20major%20h igh,early%20age%20at%20melanoma%20onset.
24. Shastry, B. S. (n.d.). SNPs: Impact on gene function and phenotype. Methods in molecular biology (Clifton, N.J.). https://pubmed.ncbi.nlm.nih.gov/19768584/#:~:text=They%20may%20influence%20promoter%20activity, and%20hence%20may%20produce%20disease
25. Skin cancer facts & statistics. The Skin Cancer Foundation. (2023, March 6). https://www.skincancer.org/skincancerinformation/skincancerfacts/
26. Stages of melanoma. AIM at Melanoma Foundation. (2023, May 9). https://www.aimatmelanoma.org/stages-of-melanoma/
27. Strachan, T., & Read, A. P. (2015). Human Molecular Genetics (4th ed.). Garland Science.
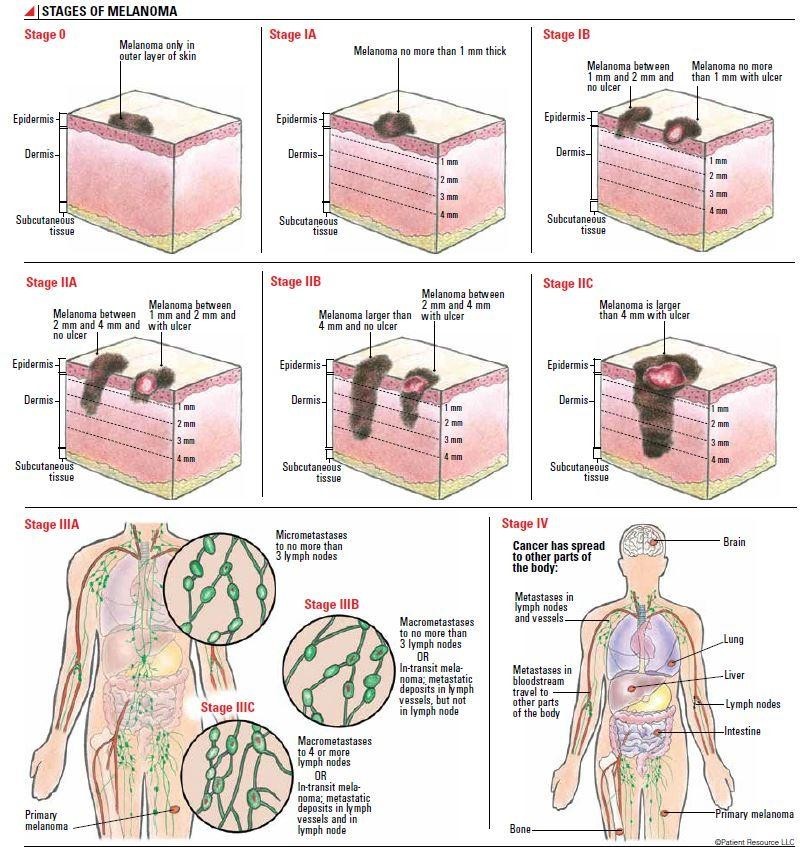
Figure 1
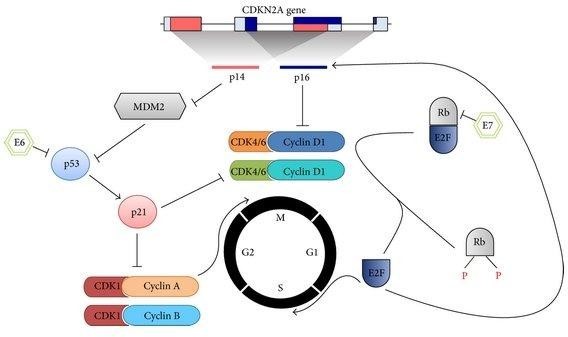
Figure 2
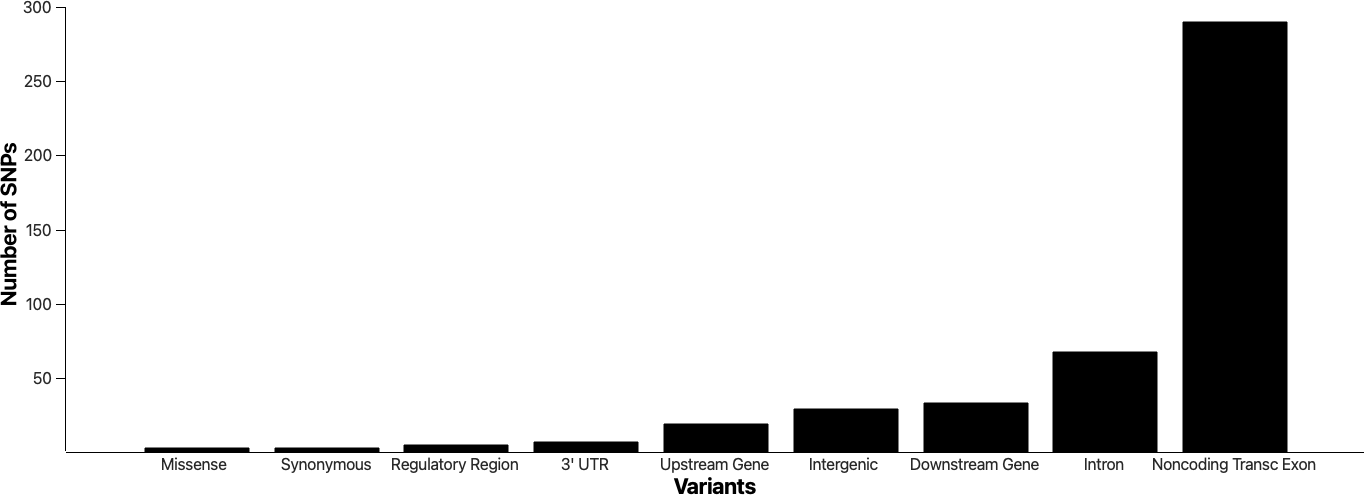
Figure 3
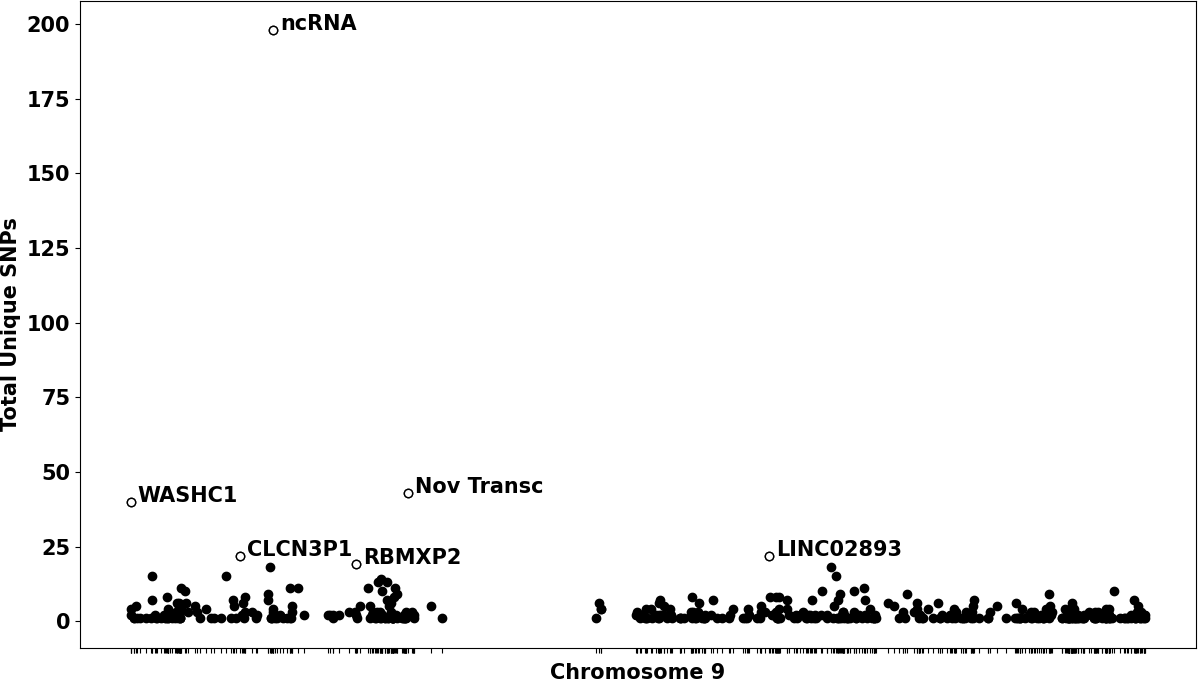
Figure 4
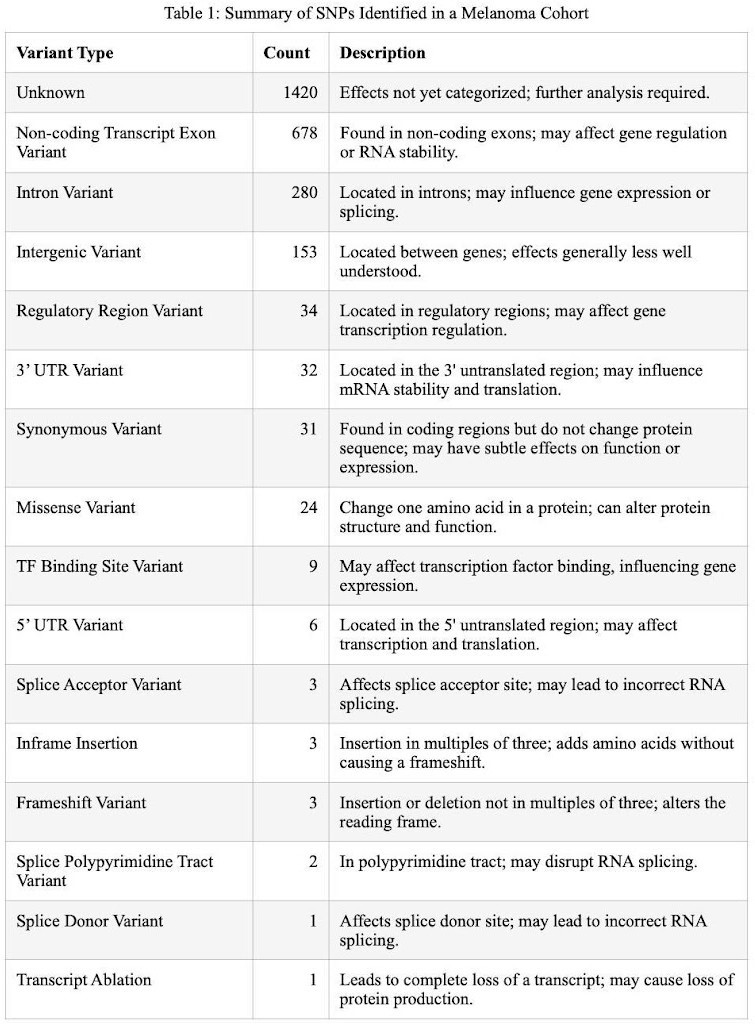
Figure 5
